Johannes Stegmaier
Juniorprofessor
105
+49 241 80 22901
Johannes.Stegmaier@lfb.rwth-aachen.de
Curriculum Vitae
| Okt 2022 – Sep 2024 |
Professurvertretung (W3) am Lehrstuhl für Bildgebung und Bildverarbeitung, RWTH Aachen |
| Apr 2018 – heute |
Juniorprofessor (W1) für biomedizinische Bildverarbeitung, RWTH Aachen |
| Okt 2016 – Jan 2017 | Postdoc am Center for Advanced Methods in Biological Image Analysis, Caltech, CA, USA |
| Feb 2015 – Dez 2017 | Postdoc am Institut für Angewandte Informatik, Karlsruher Institut für Technologie |
| Jun 2016 | Promotion (Dr.-Ing.) an der Fakultät für Maschinenbau am Karlsruher Institut für Technologie. Dissertation: “New Methods to Improve Large-Scale Microscopy Image Analysis with Prior Knowledge and Uncertainty” |
| Jan 2014 – Mai 2014 | Short Term Scholar am Janelia Research Campus, Ashburn, VA, USA |
| Feb 2012 – Jan 2015 | Wissenschaftlicher Mitarbeiter am Institut für Angewandte Informatik, Karlsruher Institut für Technologie |
| Okt 2009 – Dez 2011 | Master of Science in Bioinformatik und Systembiologie, Albert-Ludwidgs-Universität Freiburg i. Br. |
| Okt 2006 – Sep 2009 | Bachelor of Science in Bioinformatik an der Eberhard-Karls-Universität Tübingen |
Abschlussarbeiten
Veröffentlichungen
Bücher und Buchkapitel
Zebrafish: A Pharmacogenetic Model for Anesthesia
In: Methods in Enzymology

New Methods to Improve Large-Scale Microscopy Image Analysis with Prior Knowledge and Uncertainty
In: KIT Scientific Publishing
Journal-Veröffentlichungen

Denoising Diffusion Probabilistic Models for 3D Medical Image Generation
In: Scientific Reports 13 (1)

Medical Transformer for Multimodal Survival Prediction in Intensive Care: Integration of Imaging and Non-Imaging Data
In: Scientific Reports 13 (1)

Definition and Quantification of Three-Dimensional Imaging Targets to Phenotype Pre-Eclampsia Subtypes: An Exploratory Study
In: International Journal of Molecular Sciences 24 (4)

Ventral Striatal Islands of Calleja Neurons Control Grooming in Mice
In: Nature Neuroscience

MondoA Regulates Gene Expression in Cholesterol Biosynthesis-Associated Pathways Required for Zebrafish Epiboly
In: Elife 9

Pax6 Organizes the Anterior Eye Segment Independently of Optic Cup Patterning by Guiding Two Distinct Neural Crest Waves
In: PLoS Genetics 16 (6)

EmbryoMiner: A New Framework for Interactive Knowledge Discovery in Large-Scale Cell Tracking Data of Developing Embryos
In: PLOS Computational Biology 14 (4)
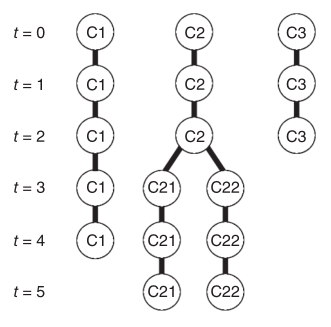
An Objective Comparison of Cell Tracking Algorithms
In: Nature Methods
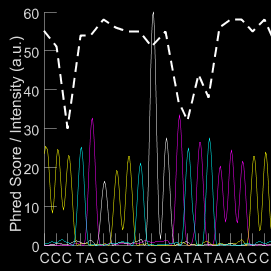
TIDE is a Simple and Effective Method to Assess Efficiency of Guide RNAs in Zebrafish
In: Zebrafish
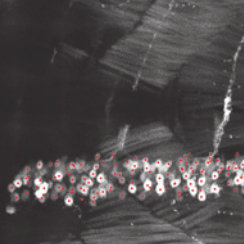
Automation Strategies for Large-Scale 3D Image Analysis
In: at-Automatisierungstechnik
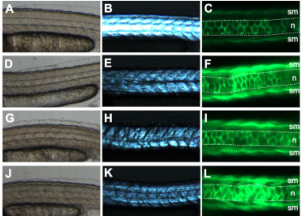
Zebrafish Biosensor for Toxicant Induced Muscle Hyperactivity
In: Scientific Reports
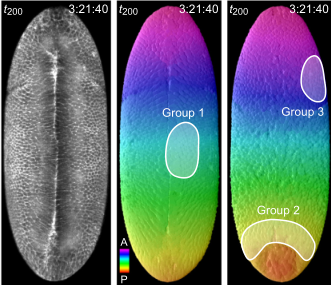
Real-Time Three-Dimensional Cell Segmentation in Large-Scale Microscopy Data of Developing Embryos
In: Developmental Cell
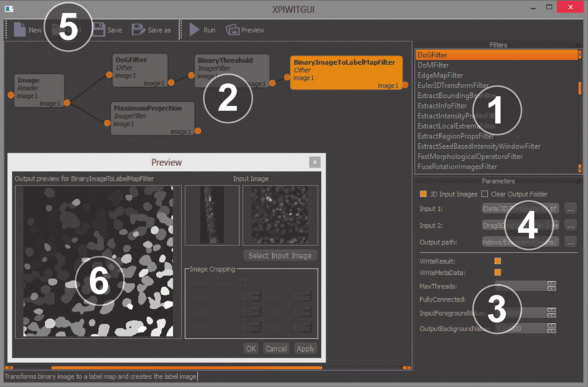
XPIWIT – An XML Pipeline Wrapper for the Insight Toolkit
In: Bioinformatics
An Ensemble-Averaged Digital Model of Zebrafish Embryo Development based on High-Speed Light-Sheet Microscopy at Single-Cell Resolution
In: Scientific Reports
Fast Segmentation of Stained Nuclei in Terabyte-Scale, Time Resolved 3D Microscopy Image Stacks
In: PLOS ONE
Automated Prior Knowledge-Based Quantification of Neuronal Patterns in the Spinal Cord of Zebrafish
In: Bioinformatics
Robust Optimal Design of Experiments for Model Discrimination using an Interactive Software Tool
In: PLOS ONE
Konferenz-Veröffentlichungen
No Free Lunch in Annotation either: An objective evaluation of Foundation models for Streamlining Annotation in Animal Tracking
In: IEEE 22nd International Symposium on Biomedical Imaging (ISBI)

Towards Annotation-Free Segmentation of Fluorescently Labeled Cell Membranes in Confocal Microscopy Images
In: MICCAI International Workshop on Simulation and Synthesis in Medical Imaging (SASHIMI)

Combined Learning for Similar Tasks with Domain-Switching Networks
In: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
Augmentations of the Bag of Visual Words Approach for Real-Time Fuzzy and Partial Image Classification
In: Proc., 27. Workshop Computational Intelligence, Dortmund, Germany

Automatic Corneal Tissue Classification Using Bag-of-Visual-Words Approaches
In: Forum Bildverarbeitung, Karlsruhe, Germany
A Framework for Feedback-based Segmentation of 3D Image Stacks
In: Current Directions in Biomedical Engineering
Generating Semi-Synthetic Validation Benchmarks for Embryomics
In: IEEE International Symposium on Biomedical Imaging (ISBI)
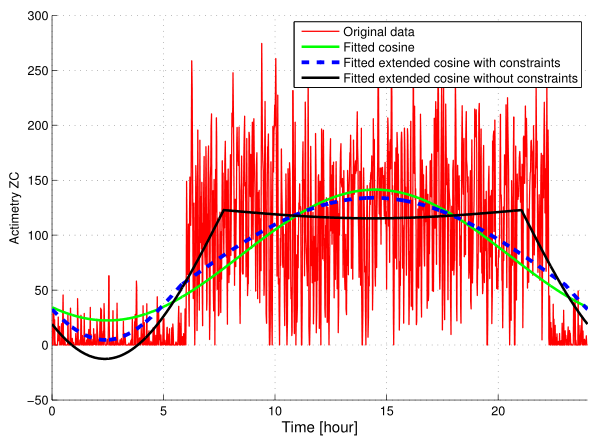
Robust Individual Circadian Parameter Estimation for Biosignal-based Personalization of Cancer Chronotherapy
In: Proc., Workshop Biosignal Processing, Berlin
Challenges of Integrating A Priori Information Efficiently in the Discovery of Spatio-Temporal Objects in Large Databases
In: Proc., 25. Workshop Computational Intelligence, Dortmund
Visualization for Error-controlled Surface Reconstruction from Large Electron Microscopy Image Stacks
In: IEEE Vis, Chicago
Challenges of Uncertainty Propagation in Image Analysis
In: Proc., 22. Workshop Computational Intelligence, Dortmund
Information Fusion of Image Analysis, Video Object Tracking, and Data Mining of Biological Images using the Open Source MATLAB Toolbox Gait-CAD
In: Biomedizinische Technik
Konferenz-Abstracts
Deep Learning Approaches to Improve Cell Segmentation and Tracking Accuracy for Interactive Knowledge Discovery in Zebrafish Embryos
In: Zebrafish Models for Human Eye Diseases, Freiburg, Germany
Interactive Cell Tracking Analysis of Light-Sheet Microscopy Data in Zebrafish Embryos using EmbryoMiner
In: 10th Anniversary Light-Sheet Fluorescence Microscopy Conference, Dresden, Germany
Automated and High-Throughput Screening Platforms for Assessing Early Zebrafish Behavior
In: 5th European Zebrafish Principal Investigator Meeting, Trento, Italy
A. Kobitski, G. U. Nienhaus, P. Sanders, U. Strähle, R. Mikut, J. Stegmaier
Interactive Knowledge Discovery in Large-Scale 3D+T Cell Tracking Data
In: EMBL Symposium ‚Seeing is Believing – Imaging the Processes of Life‘, Heidelberg, Germany
A New Framework for Interactive Knowledge-Discovery in Large-Scale Trajectory Data of Zebrafish Embryos
In: 10th European Zebrafish Meeting, Budapest, Hungary
High-Throughput Screening and Analysis of Startle Response Behavior in Zebrafish
In: 10th European Zebrafish Meeting, Budapest, Hungary
A Novel Method For Studying 3D Cellular Architecture in the Growth Plate
In: Cartilage Biology & Pathology, Lucca, Italy
Automated Approaches to Sample Handling and High-Throughput Behavioral Screening of Zebrafish
In: Zebrafish Genetics, Orlando, USA
Automated and High-Throughput Zebrafish Embryonic Behavioral Screening Platforms
In: Measuring Behavior, Dublin, Ireland
Exploiting Biological Prior Knowledge and Uncertainty for Large-Scale Image Analysis
In: EMBL Symposium ‚Seeing is Believing – Imaging the Processes of Life‘, Heidelberg, Germany
Enhancing Image Analysis Pipelines with Uncertainty Treatment
In: IEEE International Symposium on Biomedical Imaging, New York City, USA
Efficient Extraction of Cell Shapes and Nuclei from 3D Light-sheet Microscopy Images
In: BioImage Informatics, Leuven, Belgium
Embedding Biological Prior Knowledge into Efficient Processing Operators for Terabyte-Scale Image Analysis
In: EMBL Symposium ‚Seeing is Believing – Imaging the Processes of Life‘, Heidelberg, Germany
Methods for Automated Analysis of Zebrafish-related Experiments to Keep Pace with Modern High-Throughput Acquisition Technology
In: COST Workshop – Automation Methods for Zebrafish Research, Leiden, Netherlands