Digital Pathology
Motivation
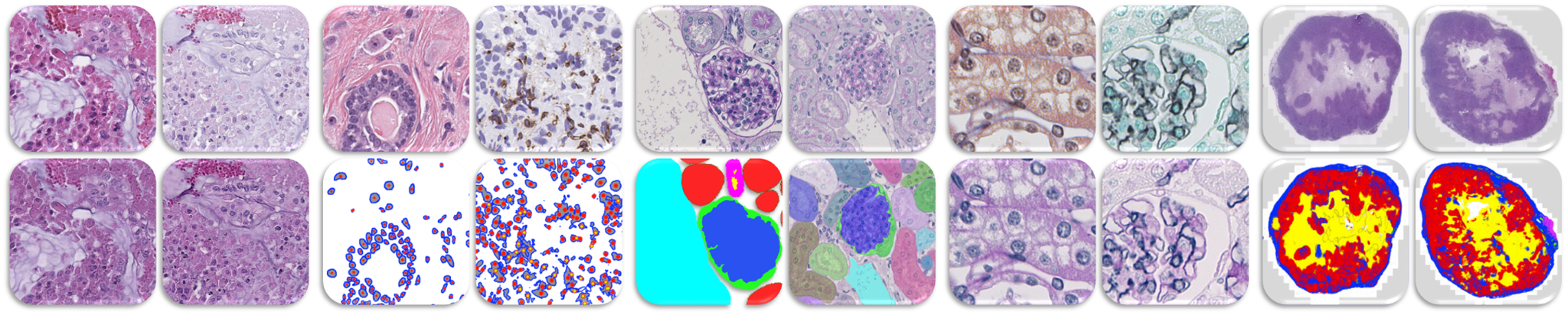
Large-scale high-throughput digitalization of histology glass slides using slide scanners, which generate digital Whole Slide Images (WSIs), and improved software and hardware solutions, together termed digital pathology, open new possibilities of automated, highly precise and reproducible quantification in pathology. In addition, digital pathology enables effective implementation of Artificial Intelligence (AI), i.e. computer systems able to perform tasks normally requiring human intelligence. Machine learning and, particularly, deep learning (DL), are research fields of AI focused on methods, in which computer systems automatically improve through experience. These approaches will most likely advance and transform pathological diagnostics in the future, leading to precise, reproducible and personalized computational pathology. Furthermore, they may even have the potential to identify new diagnostic features and biomarkers that have not been previously recognized, such as in onco-pathology.
Scientific Questions
Pathology remains an essential discipline in diagnostics of diseases, while histological analyses are key readouts of both experimental and clinical studies. At present, histopathology still relies heavily on manual and often subjective scoring, or conventional computer-assisted approaches that only allow for limited and sometimes imprecise read-outs. The state-of-the-art DL approaches focus on supervised methods which depend on training with large amounts of data labeled by experts (pathologists in case of histology). However, the acquisition of labeled medical data is often very expensive in terms of time and effort by the experts. Unsupervised methods demand large amounts of data which are, unfortunately, not always available. Moreover, there are no standardized preparation and acquisition protocols for histological images, which results in wide inter- and intra-laboratory variability of the data, such as slice thickness, stains variability, scan quality, etc. These factors greatly limit the development of DL algorithms in pathology and pathomics.
Theses
New theses are regularly advertised in the area Digital Pathology. In addition to the general overview, there are also numerous topics that have not yet been advertised, which will be gladly presented in a personal conversation.
Partners
- Prof. Dr. med. Peter Boor, Institute for Pathology, University Hospital RWTH Aachen
- Dr. rer. nat. Barbara Mara Klinkhammer, Institute for Pathology, University Hospital RWTH Aachen
- Prof. Dr. med. Friedrich Feuerhake, Institute for Pathology, Medizinische Hochschule Hannover
- Dr. Julia Schüler, Charles River Discovery Research Services Germany GmbH, Freiburg i. Br.
- Dr. Johannnes Lotz, Fraunhofer Institute for Digital Medicine MEVIS, Luebeck
- Dr. Henning Höfener, Fraunhofer Institute for Digital Medicine MEVIS, Luebeck
- Dr. André Homeyer, Fraunhofer Institute for Digital Medicine MEVIS, Luebeck
External Funding
- DFG Clinical Research Unit, “Integrating emerging methods to advance translational kidney research”, Project nr. KFO 5011
- BMBF Joint Project: “SynDICAD: Synthetic data generation for artificial intelligence application in computational biomarker analysis in digital pathology”, Project nr. FKZ: 01IS21067
- BMBF KMU-Innovativ-14 Project. “Innovative lung-cancer mouse-models recapitulating human immune response and tumor-stroma exchange (ILUMINATE)” Project nr. FKZ: 031B0006B
Contact
Publications
2022

Nassim Bouteldja, Barbara M. Klinkhammer, Tarek Schlaich, Peter Boor and Dorit Merhof
Improving Unsupervised Stain-To-Stain Translation using Self-Supervision and Meta-Learning
In: Journal of Pathology Informatics
2022

Laxmi Gupta, Barbara Mara Klinkhammer, Claudia Seikrit, Nina Fan, Nassim Bouteldja, Philipp Gräbel, Michael Gadermayr, Peter Boor and Dorit Merhof
Large-scale extraction of interpretable features provides new insights into kidney histopathology – a proof-of-concept study
In: Journal of Pathology Informatics
2021

Barbara M. Klinkhammer Nassim Bouteldja and Dorit Merhof
Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology
In: Journal of the American Society of Nephrology
2020

Daniel Bug, Gregor Nickel, Anne Grote, Friedrich Feuerhake, Eva Oswald, Julia Schüler and Dorit Merhof
Image Quilting for Histological Image Synthesis
In: Bildverarbeitung für die Medizin (BVM)
2020

Daniel Bug, Felix Bartsch, Nadine Sarah Schaadt, Mathias Wien, Friedrich Feuerhake, Julia Schüler, Eva Oswald and Dorit Merhof
Scalable HEVC for Histological Whole-Slide Image Compression
In: Bildverarbeitung für die Medizin (BVM)
2019

Michael Gadermayr, Ann-Kathrin Dombrowski, Barbara Mara Klinkhammer, Peter Boor and Dorit Merhof
CNN Cascades for Segmenting Sparse Objects in Gigapixel Whole Slide Images
In: Computerized Medical Imaging and Graphics 71
2019

Michael Gadermayr, Laxmi Gupta, Vitus Appel, Peter Boor, Barbara Mara Klinkhammer and Dorit Merhof
Generative Adversarial Networks for Facilitating Stain-Independent Supervised & Unsupervised Segmentation: A Study on Kidney Histology
In: IEEE Transactions on Medical Imaging
2019

Michael Gadermayr, Laxmi Gupta, Barbara Mara Klinkhammer, Peter Boor and Dorit Merhof
Unsupervisedly Training GANs for Segmenting Digital Pathology with Automatically Generated Annotations
In: 2nd International Conference on Medical Imaging with Deep Learning (MIDL)
2019

Daniel Bug, Friedrich Feuerhake, Eva Oswald, Julia Schüler and Dorit Merhof
Semi-Automated Analysis of Digital Whole Slides from Humanized Lung-Cancer Xenograft Models for Checkpoint Inhibitor Response Prediction
In: Oncotarget
2019

Daniel Bug, Dennis Eschweiler, Qianyu Liu, Justus Schock, Leon Weninger, Friedrich Feuerhake, Julia Schüler, Johannes Stegmaier and Dorit Merhof
Combined Learning for Similar Tasks with Domain-Switching Networks
In: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
2019

Philipp Gräbel Daniel Bug and Dorit Merhof
Supervised and Unsupervised Cell-Nuclei Detection in Immunohistology
In: 2nd MICCAI Workshop on Computational Pathology (COMPAY)
2019

Laxmi Gupta, Barbara Mara Klinkhammer, Peter Boor, Dorit Merhof and Michael Gadermayr
GAN-Based Image Enrichment in Digital Pathology Boosts Segmentation Accuracy
In: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
2019

Laxmi Gupta, Barbara Mara Klinkhammer, Peter Boor, Dorit Merhof and Michael Gadermayr
Iterative learning to make the most of unlabeled and quickly obtained labeled data in histology
In: 2nd International Conference on Medical Imaging with Deep Learning (MIDL)
2018

Laxmi Gupta, Barbara Mara Klinkhammer, Peter Boor, Dorit Merhof and Michael Gadermayr
Stain Independent Segmentation of Whole Slide Images: A Case Study in Renal Histology
In: IEEE International Symposium on Biomedical Imaging (ISBI)
2018

Michael Gadermayr, Dennis Eschweiler, Barbara Mara Klinkhammer, Peter Boor and Dorit Merhof
Gradual Domain Adaptation for Segmenting Whole Slide Images Showing Pathological Variability
In: International Conference on Image and Signal Processing (ICISP)
2018

Michael Gadermayr, Vitus Appel, Barbara M. Klinkhammer, Peter Boor and Dorit Merhof
Which Way Round? A Study on the Performance of Stain-Translation for Segmenting Arbitrarily Dyed Histological Images
In: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)
2017

Michael Gadermayr, Sean Steven Cooper, Barbara Klinkhammer, Peter Boor and Dorit Merhof
A Quantitative Assessment of Image Normalization for Classifying Histopathological Tissue of the Kidney
In: German Conference on Pattern Recognition (GCPR)
2017

Michael Gadermayr, Dennis Eschweiler, Abiramjee Jeevanesan, Barbara Mara Klinkhammer, Peter Boor and Dorit Merhof
Segmenting Renal Whole Slide Images Virtually Without Training Data
In: Computers in Biology and Medicine 90
2017

Daniel Bug, Anne Grote, Julia Schüler, Friedrich Feuerhake and Dorit Merhof
Analyzing Immunohistochemically Stained Whole-Slide Images of Ovarian Carcinoma
In: Bildverarbeitung für die Medizin (BVM)
2017

Daniel Bug, Steffen Schneider, Anne Grote, Eva Oswald, Friedrich Feuerhake, Julia Schüler and Dorit Merhof
Context-based Normalization of Histological Stains using Deep Convolutional Features
In: MICCAI International Workshop on Deep Learning in Medical Image Analysis (DLMIA)
2016

Michael Gadermayr, Barbara M. Klinkhammer, Peter Boor and Dorit Merhof
Do we Need Large Annotated Training Data for Detection Applications in Biomedical Image Data? A Case Study in Renal Glomeruli Detection
In: MICCAI Workshop on Machine Learning in Medical Imaging (MLMI)
2016

Michael Gadermayr, Martin Strauch, Barbara Mara Klinkhammer, Sonja Djudjaj, Peter Boor and Dorit Merhof
Domain Adaptive Classification for Compensating Variability in Histopathological Whole Slide Images
In: International Conference on Image Analysis and Recognition (ICIAR)
2016

Daniel Bug, Julia Schüler, Friedrich Feuerhake and Dorit Merhof
Multi-class single-label classification of histopathological whole-slide images
In: IEEE International Symposium on Biomedical Imaging (ISBI)
2015

Daniel Bug, Friedrich Feuerhake and Dorit Merhof
Foreground Extraction for Histopathological Whole-Slide Imaging
In: Bildverarbeitung für die Medizin (BVM)