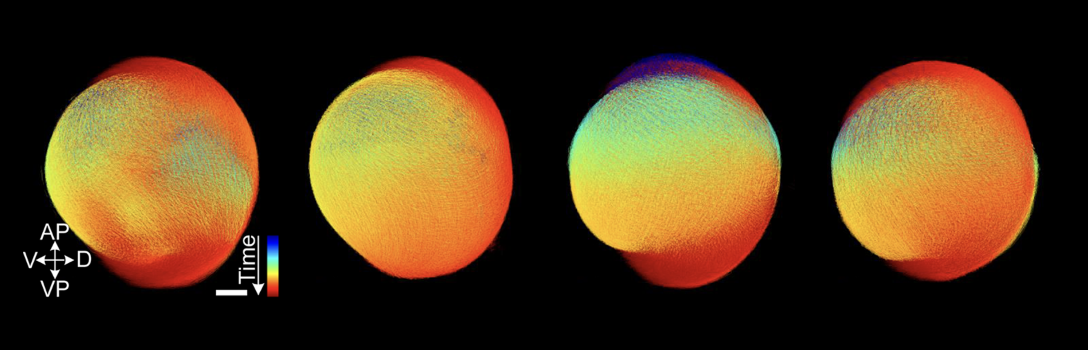
Automatic Spatiotemporal Alignment of Large-Scale 3D+t Point Clouds
Motivation
Multidimensional fluorescence microscopy has become a common technique in biology labs all over the world as it represents an invaluable tool to study early embryonic development in space and time (3D+t) at cellular resolution. Automatic segmentation and tracking algorithms are used to extract thousands of cell movement trajectories from potentially terabyte-scale 3D+t image data sets that offer the possibility for a detailed analysis of inter-individual differences. A fundamental problem that remains after having obtained such tracked point clouds, however, is the comparison of individual experiments to confirm biological hypotheses in multiple repeats. The lack of fully automated solutions to this 3D+t alignment problem currently limits whole-embryo analyses to simple specimens, early time points or manual analyses.
Scientific Questions
The aim of this project is the development of new methods for automated spatiotemporal alignment of large 3D+t point clouds. As complex organisms usually lack one-to-one cell correspondences that could be used for registration, a fundamental part of the project will be the development of generic descriptors to identify various anatomical regions at different developmental stages using both classical and machine learning-based approaches. These descriptors will then be used to obtain a spatiotemporal registration of the point clouds. Moreover, we generate synthetic training data for the machine learning approaches using a comprehensive simulation platform that allows mimicking embryonic development of different specimens at multiple levels of detail.
Theses
New theses are regularly advertised in the area of automatic processing of large-scale point cloud data. In addition to the general overview there are also numerous topics that have not yet been advertised, which will be gladly presented in a personal conversation.
Partners
- G. U. Nienhaus and Dr. A. Kobitski, Institute of Applied Physics, Karlsruhe Institute of Technology (KIT)
- Prof. U. Strähle and Dr. M. Takamiya, Institute of Biological and Chemical Systems – Biological Information Processing,
- Karlsruhe Institute of Technology (KIT)
- apl. Prof. R. Mikut, Institute for Automation and Applied Informatics, Karlsruhe Institute of Technology (KIT)
External Funding
- DFG Research Grant, “Automatic spatiotemporal alignment of large-scale 3D+t point clouds”, Projektnummer 432051322
Contact
Publications

Surrounding Cell Suppression for Unsupervised Representation Learning in Hematological Cell Classification
In: IEEE International Symposium on Biomedical Imaging (ISBI)
Automatic Spatiotemporal Alignment of Large-Scale 3D+t Point Clouds
Motivation
Multidimensional fluorescence microscopy has become a common technique in biology labs all over the world as it represents an invaluable tool to study early embryonic development in space and time (3D+t) at cellular resolution. Automatic segmentation and tracking algorithms are used to extract thousands of cell movement trajectories from potentially terabyte-scale 3D+t image data sets that offer the possibility for a detailed analysis of inter-individual differences. A fundamental problem that remains after having obtained such tracked point clouds, however, is the comparison of individual experiments to confirm biological hypotheses in multiple repeats. The lack of fully automated solutions to this 3D+t alignment problem currently limits whole-embryo analyses to simple specimens, early time points or manual analyses.
Scientific Questions
The aim of this project is the development of new methods for automated spatiotemporal alignment of large 3D+t point clouds. As complex organisms usually lack one-to-one cell correspondences that could be used for registration, a fundamental part of the project will be the development of generic descriptors to identify various anatomical regions at different developmental stages using both classical and machine learning-based approaches. These descriptors will then be used to obtain a spatiotemporal registration of the point clouds. Moreover, we generate synthetic training data for the machine learning approaches using a comprehensive simulation platform that allows mimicking embryonic development of different specimens at multiple levels of detail.
Theses
New theses are regularly advertised in the area of automatic processing of large-scale point cloud data. In addition to the general overview there are also numerous topics that have not yet been advertised, which will be gladly presented in a personal conversation.
Partners
- G. U. Nienhaus and Dr. A. Kobitski, Institute of Applied Physics, Karlsruhe Institute of Technology (KIT)
- Prof. U. Strähle and Dr. M. Takamiya, Institute of Biological and Chemical Systems – Biological Information Processing,
- Karlsruhe Institute of Technology (KIT)
- apl. Prof. R. Mikut, Institute for Automation and Applied Informatics, Karlsruhe Institute of Technology (KIT)
External Funding
- DFG Research Grant, “Automatic spatiotemporal alignment of large-scale 3D+t point clouds”, Projektnummer 432051322
Contact
Publications

Surrounding Cell Suppression for Unsupervised Representation Learning in Hematological Cell Classification
In: IEEE International Symposium on Biomedical Imaging (ISBI)