Synthesis, Segmentation, and Tracking of Live Cell Microscopy Image Data
Motivation
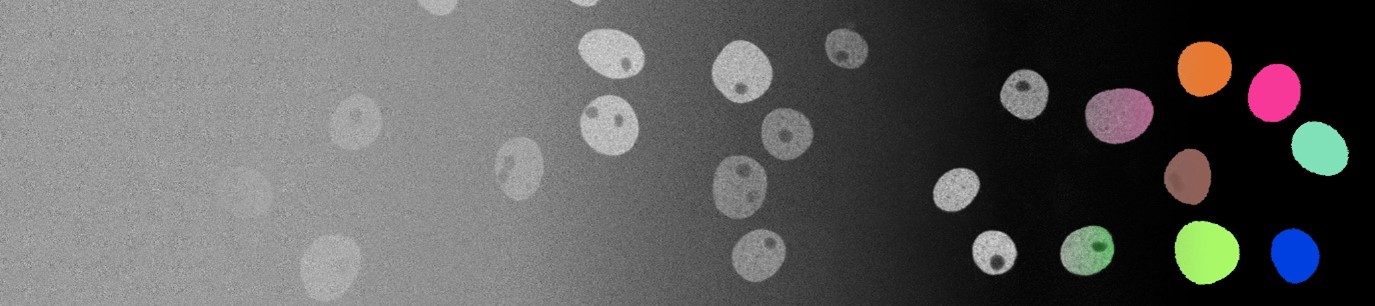
Developmental biology focuses on studying the processes of how cells migrate, interact and divide. Specifically, cell location and morphology, analyzed by segmentation and tracking approaches, constitute one of the key tasks in understanding the underlying principles of development. Technological advances allow biologists to capture and store extensive amounts of highly detailed microscopy image data, which renders the application of automated approaches essential. However, the potential of recent deep learning-based approaches is severely limited by the scarcity of fully annotated image data sets. Therefore, this project focuses on synthesis of fully annotated microscopy video data sets, eliminating the need for time-consuming manual annotation efforts.
Scientific Questions
- How to generate realistic fully synthetic cell textures and shapes based on a limited amount of annotated real data?
- How to capture the spatiotemporal relations of cell bodies and reflect this on the generated data?
- How to make sure that there is enough variability in the synthetic data set and assess its quality?
- How much improvement can we get by training segmentation and tracking models on abundant synthetic data as opposed to training them with limited real data?
Partners
- G. U. Nienhaus and Dr. A. Kobitski, Institute of Applied Physics, Karlsruhe Institute of Technology (KIT)
- Prof. U. Strähle and Dr. M. Takamiya, Institute of Biological and Chemical Systems – Biological Information Processing, Karlsruhe Institute of Technology (KIT)
- apl. Prof. R. Mikut, Institute for Automation and Applied Informatics, Karlsruhe Institute of Technology (KIT)
External Funding
- DFG Research Grant, “On-the-fly Datensynthese für eine auf Deep Learning basierende Analyse von 3D+t Mikroskopie Experimenten”, Project Nr. 447699143
Contact
Publications

Towards Annotation-Free Segmentation of Fluorescently Labeled Cell Membranes in Confocal Microscopy Images
In: MICCAI International Workshop on Simulation and Synthesis in Medical Imaging (SASHIMI)











